Chapter 1 Data loading and cleaning
The objective of exploratory data anlysis is to understand your data, find patterns, identify outliers, and possibly form hypotheses for further analysis. In this chapter, we will use the penguins dataset to learn how to load data, inspect it, and process it by removing missing values and outliers. The following Chapter 2 will cover more details on data transformation. Chapter 3 will cover data visualization.
Let’s start with loading the tidyverse packages:
1.1 Load data
We use the function readr::read_csv to load the data. This function and its variations like readr::read_delim are able to read a wide variety of formats. Here, we read a compressed file in comma separated (csv) format.
## Rows: 347 Columns: 7
## ── Column specification ───────────────────────────────────────────────────────────────────────
## Delimiter: ","
## chr (3): species, island, sex
## dbl (4): bill_length_mm, bill_depth_mm, flipper_length_mm, body_mass_g
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.We learn that the data set contains 347 observations with 7 columns.1 Three of the columns are string (chr: species, island, sex), the rest are numeric (dbl: bill_length_mm, bill_depth_mm, flipper_length_mm, body_mass_g).
Note that the command read_csv is spelt similarly to the base-R function read.csv. To avoid confusion, we use the notation readr::read_csv to indicate that we use the function read_csv from the package readr.
Todo:
Read the help page for readr::read_csv and find out what the arguments file, delim, comment, and skip do.
1.2 Inspect data to identify missing values
The data is now stored in the variable penguins as a tibble. A tibble is the tidyverse form of a data frame. It is a bit more strict than a data frame, but also more consistent. Let’s have a look at the first few rows of the data:
## # A tibble: 347 × 7
## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
## <chr> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 Adelie Torgersen 39.1 18.7 181 3750
## 2 Adelie Torgersen 39.5 17.4 186 3800
## 3 Adelie Torgersen 40.3 18 195 3250
## 4 Adelie Torgersen NA NA NA NA
## 5 Adelie Torgersen 36.7 19.3 193 3450
## 6 Adelie Torgersen 39.3 20.6 190 3650
## 7 Adelie Torgersen 38.9 17.8 181 3625
## 8 Adelie Torgersen 39.2 19.6 195 4675
## 9 Adelie Torgersen 34.1 18.1 193 3475
## 10 Adelie Torgersen 42 20.2 190 4250
## # ℹ 337 more rows
## # ℹ 1 more variable: sex <chr>Compared to standard data frames, this is a more informative representation of your data. It shows the first few rows, the column names, and the data types of the columns. The data types are important, because they determine what you can do with the data.
A useful first step is to check for missing data. We can already see in the output above, that the dataset contains missing values indicated by NA. There are various ways of doing this. One way is to use the function is.na to check for missing values in each column:
## species island bill_length_mm bill_depth_mm
## 0 0 2 2
## flipper_length_mm body_mass_g sex
## 2 2 11Several columns contain missing values. Here are the rows with missing values:
## # A tibble: 11 × 7
## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
## <chr> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 Adelie Torgersen NA NA NA NA
## 2 Adelie Torgersen 34.1 18.1 193 3475
## 3 Adelie Torgersen 42 20.2 190 4250
## 4 Adelie Torgersen 37.8 17.1 186 3300
## 5 Adelie Torgersen 37.8 17.3 180 3700
## 6 Adelie Dream 37.5 18.9 179 2975
## 7 Gentoo Biscoe 44.5 14.3 216 4100
## 8 Gentoo Biscoe 46.2 14.4 214 4650
## 9 Gentoo Biscoe 47.3 13.8 216 4725
## 10 Gentoo Biscoe 44.5 15.7 217 4875
## 11 Gentoo Biscoe NA NA NA NA
## # ℹ 1 more variable: sex <chr>With just a few missing values in the dataset, we can simply remove them. This is done with the function drop_na:
The pipe operator %>% (pronounced as then) is used to chain commands together. The command penguins %>% drop_na() takes the data frame penguins and passes it to the function drop_na. The statement is equivalent to:
df <- drop_na(penguins)The real power of the pipe operator becomes apparent when we combined multiple commands in a processing pipeline.
1.3 Identifying outliers
The tidyverse comes with a powerful plotting package called ggplot2. We will use the ggplot2 package throughout this class. To get started, we look at the values of the numerical columns to get a feel for the data. Here is a graph (Figure 1.1 of the values of the bill_length_mm column:
Code
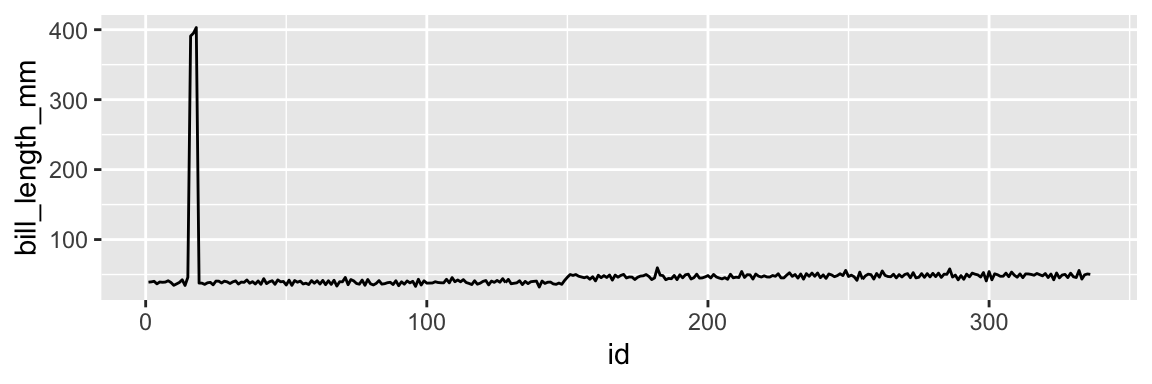
Figure 1.1: Values of the bill_length_mm column.
We first create a temporary tibble df that contains the id column and the bill_length_mm column. The id column is just a sequence of numbers from 1 to 347. We use this column to plot the values of bill_length_mm against the row number. The geom_line function adds a line plot to the plot. The result is a line plot of the values of bill_length_mm against the row number. We can see that the values are mostly between 30 and 60, but there are some outliers. The values are:
| id | species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex |
|---|---|---|---|---|---|---|---|
| 19 | Adelie | Torgersen | 34.4 | 18.4 | 184 | 3325 | female |
| 20 | Adelie | Torgersen | 46.0 | 21.5 | 194 | 4200 | male |
| 21 | Adelie | Torgersen | 391.0 | 18.7 | 181 | 3750 | male |
| 22 | Adelie | Torgersen | 395.0 | 17.4 | 186 | 3800 | female |
| 23 | Adelie | Torgersen | 403.0 | 18.0 | 195 | 3250 | female |
| 24 | Adelie | Biscoe | 37.8 | 18.3 | 174 | 3400 | female |
| 25 | Adelie | Biscoe | 37.7 | 18.7 | 180 | 3600 | male |
We can see that rows 21, 22, and 23 have values that are around 10 times higher than all other values. It’s likely that these values are due to an error in the data collection process. We can remove these values with the following command:
Todo:
Check if any of the other numerical columns contain outliers.
1.4 Summary
In this chapter, we have learned how to load data, inspect it, and process it. We have also learned how to remove missing values and outliers.
The analysis of this chapter results in the following processing pipeline:
Code
## Rows: 347 Columns: 7
## ── Column specification ───────────────────────────────────────────────────────────────────────
## Delimiter: ","
## chr (3): species, island, sex
## dbl (4): bill_length_mm, bill_depth_mm, flipper_length_mm, body_mass_g
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.Further information:
The data import cheatsheet is a short summary of all the main features of readr and readxl.
For more details see https://readr.tidyverse.org/.
The readxl package allows to import Excel files, see https://readxl.tidyverse.org/.
Code
The code of this chapter is summarized here.
Code
knitr::opts_chunk$set(echo=TRUE, cache=TRUE, autodep=TRUE, fig.align="center")
library(tidyverse)
penguins <- readr::read_csv("data/penguins_modified.csv.gz")
penguins
colSums(is.na(penguins))
penguins[rowSums(is.na(penguins)) > 0, ]
df <- penguins %>%
drop_na()
df <- penguins %>%
drop_na() %>%
mutate(id = row_number())
ggplot(df, aes(x=id, y=bill_length_mm)) + geom_line()
penguins %>%
mutate(id = row_number(), .before=species) %>%
slice(19:25) %>%
knitr::kable()
df <- penguins %>%
drop_na() %>%
filter(bill_length_mm < 100)
penguins <- readr::read_csv("data/penguins_modified.csv.gz") %>%
drop_na() %>%
filter(bill_length_mm < 100)Note: we use a modified version of the original dataset here↩︎